Structural plasticity mediates distinct GAP‐dependent GTP hydrolysis mechanisms in Rab33 and Rab5 - Majumdar - 2017 - The FEBS Journal - Wiley Online Library
Energy profile and chemical transformations upon GTP hydrolysis in the... | Download Scientific Diagram

Inhibition and Termination of Physiological Responses by GTPase Activating Proteins | Physiological Reviews

See previous page) (A) rates of intrinsic and GAP-mediated hydrolysis... | Download Scientific Diagram

Phospholipase C-β1 Directly Accelerates GTP Hydrolysis by Gαq and Acceleration Is Inhibited by Gβγ Subunits* - Journal of Biological Chemistry

Physicochemical Insights into Microscopic Events Driven by GTP Hydrolysis Reaction in Ras-GAP system | bioRxiv

Molecules | Free Full-Text | Structural Insights into the Regulation Mechanism of Small GTPases by GEFs | HTML

Rab Proteins Regulate Membrane Fusion Through GTP Hydrolysis. (Left)... | Download Scientific Diagram

GTP Hydrolysis Is Not Important for Ypt1 GTPase Function in Vesicular Transport | Molecular and Cellular Biology

Ras and GTPase-activating protein (GAP) drive GTP into a precatalytic state as revealed by combining FTIR and biomolecular simulations | PNAS

Figure 3 from Hydrolysis of Guanosine Triphosphate (GTP) by the Ras·GAP Protein Complex: Reaction Mechanism and Kinetic Scheme. | Semantic Scholar
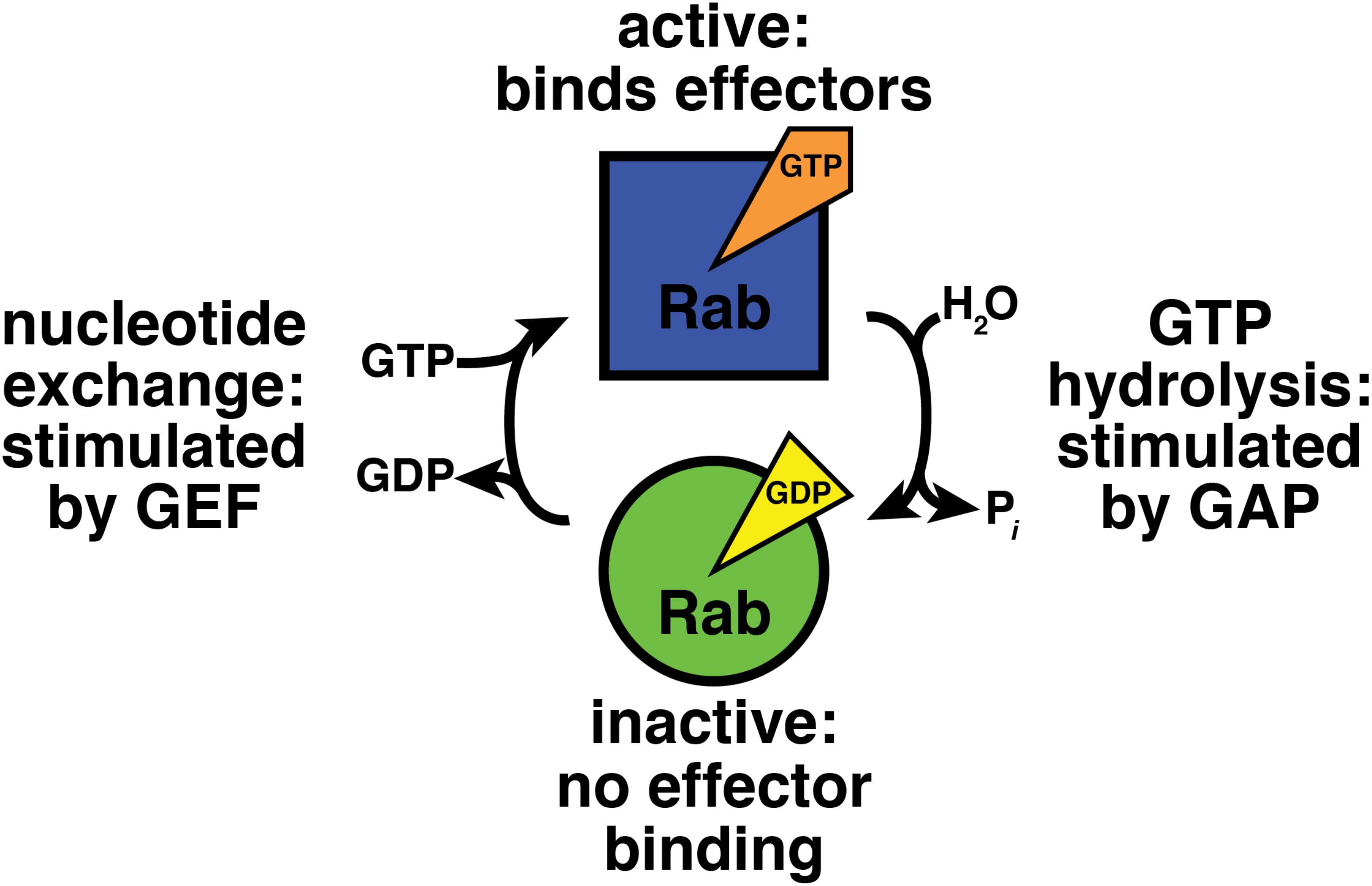
Frontiers | This Is the End: Regulation of Rab7 Nucleotide Binding in Endolysosomal Trafficking and Autophagy | Cell and Developmental Biology

Diversity of mechanisms in Ras–GAP catalysis of guanosine triphosphate hydrolysis revealed by molecular modeling - Organic & Biomolecular Chemistry (RSC Publishing) DOI:10.1039/C9OB00463G
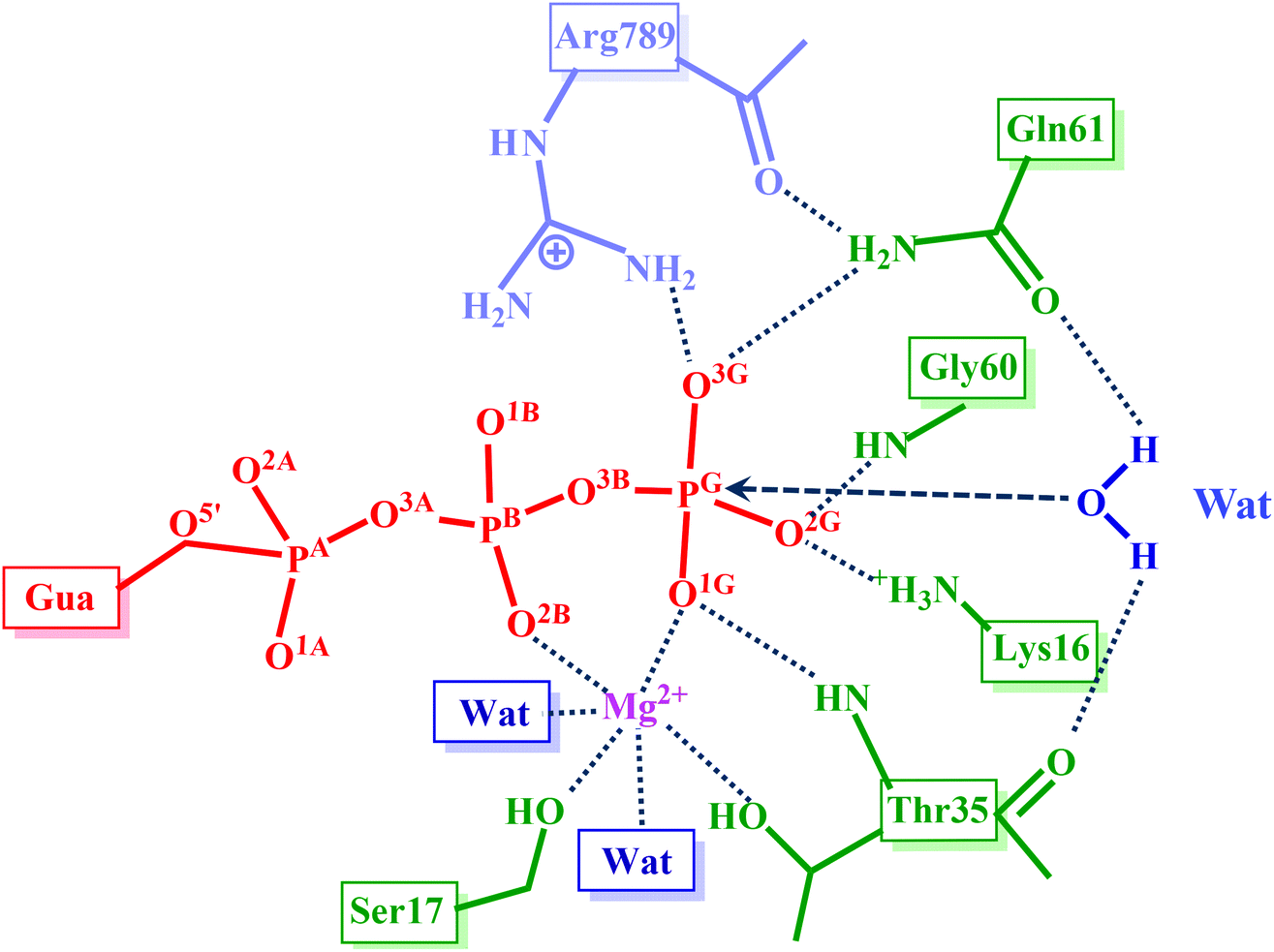
Diversity of mechanisms in Ras–GAP catalysis of guanosine triphosphate hydrolysis revealed by molecular modeling - Organic & Biomolecular Chemistry (RSC Publishing) DOI:10.1039/C9OB00463G

The GTPase-activating Protein RGS4 Stabilizes the Transition State for Nucleotide Hydrolysis* - Journal of Biological Chemistry

The pre-hydrolysis state of p21ras in complex with GTP: new insights into the role of water molecules in the GTP hydrolysis reaction of ras-like proteins: Structure



![PDF] Modeling the mechanisms of biological GTP hydrolysis. | Semantic Scholar PDF] Modeling the mechanisms of biological GTP hydrolysis. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/d73a3c179ccd59a0d10afd8fcec901434302c963/2-Figure1-1.png)